Wetlab
Fungus Identification
Overview:
WHY: The initial goal of our project was to degrade the fungi present on cellulose-rich building materials. We assumed Stachybotrys to be the primary mold fungi but consequently discovered Aspergillus, Alternaria, Penicillium, and Rhizopus commonly found on walls. With this motivation, we started sampling the fungi molds present on walls in our institution.
HOW IT WILL HELP: Sampling and identifying fungi in our institutional campus enabled us to do in-house antifungal assays and early growth modeling.
SPECIES IDENTIFIED: We identified around seven different species of fungi on the campus
Process of Identification
- Collection of samples: All fungus species used for the project were natively collected from the IISER Thiruvananthapuram campus. The molds were spotted on walls and rooftops and were collected by wearing gloves and scraping the surface spatula, following appropriate safety protocols, and transferred to PDA plates.
- Culturing: After the samples were isolated from the environment, they were transferred to potato dextrose agar plates. Such isolations consisted of multiple species of fungi, and single colonies were further isolated into new PDA plates through inoculums. To reduce the flow of spores inside the biosafety cabinet while working on the fungi, we started culturing the species in slant PDA as suggested by Dr.Hariprasad P. After isolating single species in slant, we proceeded towards identification based on (a) morphology (b) genomic DNA sequencing. The plates were routinely subcultured to preserve the fungi species. The fungi were placed in 40% glycerol stock and maintained at -80C. The media were incubated at 28 degrees celsius in an incubator. To eliminate contaminants in the culture, ampicillin was added to the media.
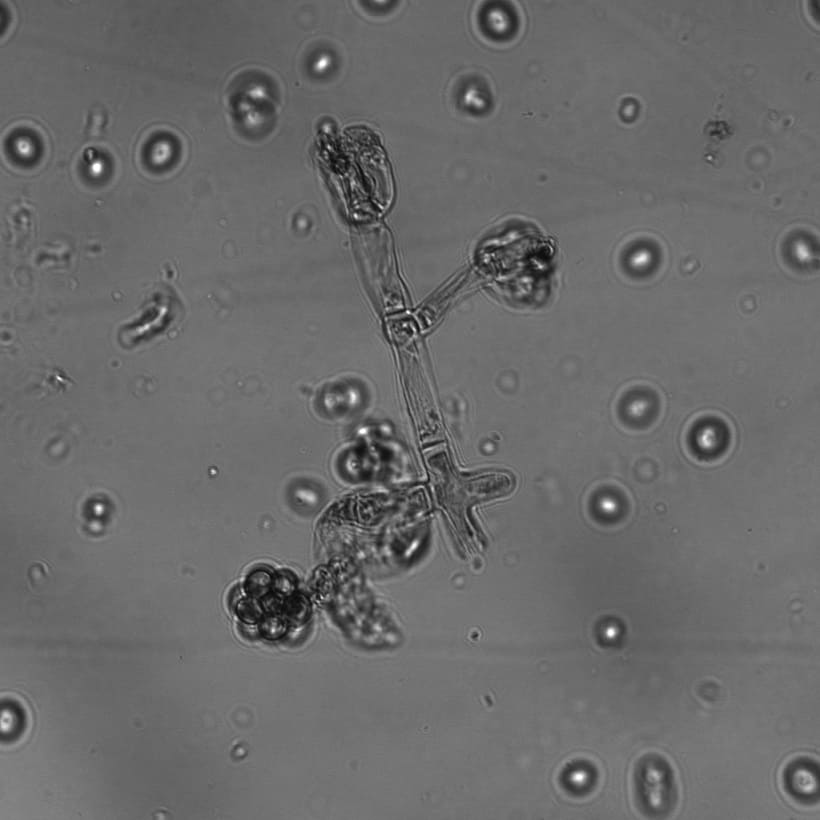
- Morphology-based identification: Certain species of fungi have characteristic morphological features viz. colony color variation, hyphae and spore structure and arrangement, growth rate. Fungus slides were made stained with lactophenol cotton blue and observed under Brightfield, Confocal, Differential Interference Contrast (DIC), and Scanning Electron Microscope (SEM). Predictions made based on morphology were further confirmed through sequencing. [add photos here]
- DNA isolation: For extraction of DNA, the fungal cultures were first inoculated in PDB media in conical flasks. Post mycelial growth in the media, the mycelium beads were harvested from the culture broth and filtered through filter paper. They were then subsequently freeze-dried in liquid nitrogen, and DNA was isolated through the CTAB method. The genomic DNA was PCR amplified using (Internal Transcribed Spacer region) ITS 4, and ITS 5 primer Agarose Gel Electrophoresis confirmed the amplification. To eliminate residual nucleotides, primers, and buffer components, PCR clean-up was performed before sequencing.
- Sequencing: The Sanger pre-sequencing consists of (a) Sanger PCR for linear amplification of our gene (b) PCR clean up using ethanol precipitation method. DNA Sanger sequencing by means of capillary electrophoresis was performed using the Thermofisher 3500 ABI genetic analyzer at the IISER TVM Genomics lab facility. The sequence electropherogram was verified to contain the necessary data. Electropherogram data was then subsequently converted to FASTA sequences and sequence aligned using NCBI Nucleotide BLAST to identify species. The species identified were: Rhizopus oryzae, Phanerochaete sodida, Fusarium solani , Nodulisporium indicum, Trichoderma, Aspergillus versicolor.